 |
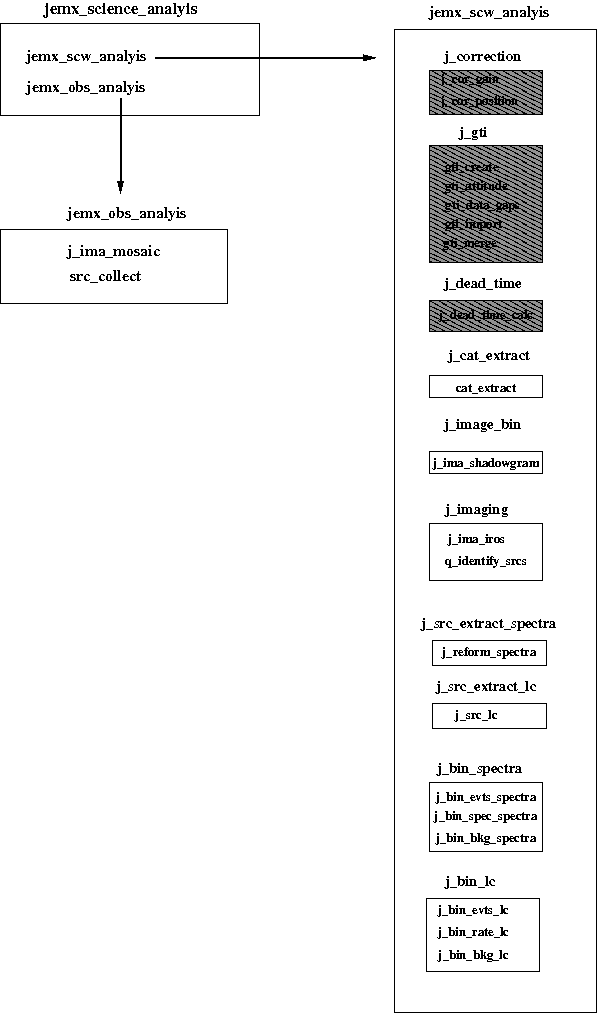
In the previous cookbook chapter (Section 6) several examples of the JEM-X data scientific analysis along with the description of the results were given. There you have seen that in order to run Scientific Analysis you should just launch the main script jemx_science_analysis with a desired set of parameters. As it was discussed in the Overview (Section 5) processing of the main script consists mainly of a loop over the pointings in the Observation Group calling the script jemx_scw_analysis, which in turn consists of smaller scripts unifying the executables with the similar tasks, see Figure 22. In the present chapter we describe these small scripts in more details in order to explain how the main script works and what parameters you have to enter for a proper analysis.
Describing the executables we mention all the parameters that were included as a parameter to the main script. All other parameters are set internally. To know about them type the name of the desired executable with -v option. Non-hidden parameters of the main script are marked with a bold font. Usually names of the main script parameters are derived from the corresponding name of the executable parameters by adding as prefix the name of the OSA level at which they are called (e.g. parameter diameter of the executable j_ima_mosaic called at IMA2 level of OSA, is named IMA2_diameter in the main script. In the rare case when this rule is broken we give both names in the description of the executable. The detailed description of the results produced at each step can be found in the Appendix section.
In the Appendix you also find the description of raw and prepared data with which you start the analysis.
 |